Markov Chain Monte Carlo Using PYMC¶
This example demonstrates Markov Chain Monte Carlo using PYMC (https://pymc-devs.github.io/pymc/) with the MCMC function.
%matplotlib inline
import sys,os
from numpy import array, double, arange, random
try:
import matk
except:
try:
sys.path.append(os.path.join('..','src'))
import matk
except ImportError as err:
print 'Unable to load MATK module: '+str(err)
# Define basic function
def f(pars):
a = pars['a']
c = pars['c']
m=double(arange(20))
m=a*(m**2)+c
return m
# Create matk object
prob = matk.matk(model=f)
# Add parameters with 'true' parameters
prob.add_par('a', min=0, max=10, value=2)
prob.add_par('c', min=0, max=30, value=5)
# Run model using 'true' parameters
prob.forward()
# Create 'true' observations with zero mean, 0.5 st. dev. gaussian noise added
prob.obsvalues = prob.simvalues + random.normal(0,0.1,len(prob.simvalues))
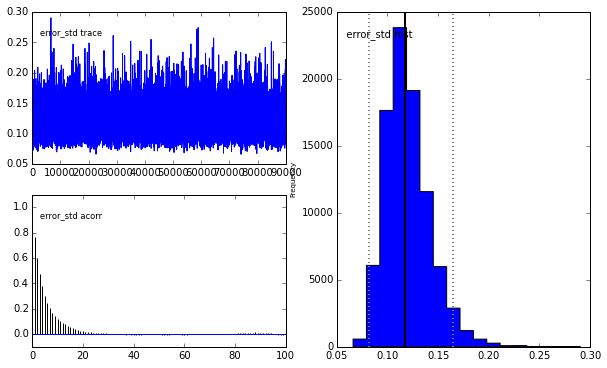
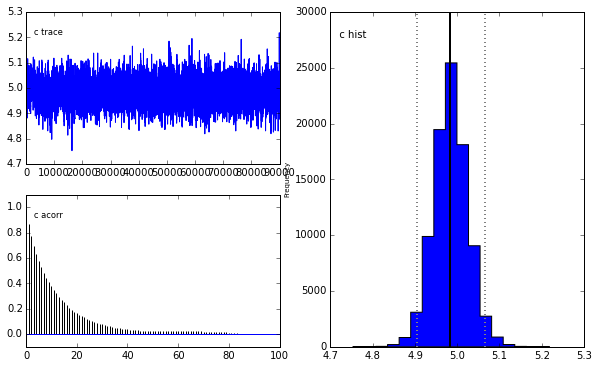
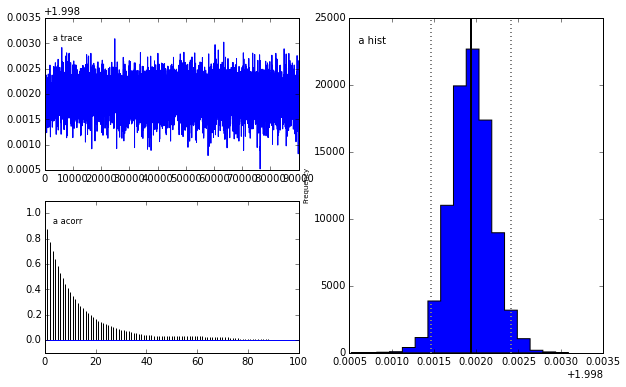
# Run MCMC with 100000 samples burning (discarding) the first 10000
M = prob.MCMC(nruns=100000,burn=10000,verbose=False)
[-----------------100%-----------------] 100000 of 100000 complete in 128.1 sec
# Plot results, PNG files will be created in current directory
# It is apparent that the true parameter values and standard deviation
# are recovered as the most likely values.
prob.MCMCplot(M)
Plotting error_std
Plotting c
Plotting a